Code
# data loading, manipulation, and writing
library(tidyverse)A quick guide to calculating 24-motion variables
March 20, 2018
March 24, 2019
This is a quick guide to calculate the 24 motion parameters from the original six motion parameters. These 24 motion parameters can be used as control variables (covariates of no interest; nuisance regressors) that is typically performed in fMRI first-level analyses, especially in resting-state and task-based functional connectivity analyses.
A helper function to plot the motion parameters.
# graph motion parameters and facet by motion type
graph_motion <- function(dataset, title) {
require(reshape2)
data_long <- melt(dataset, id.var = "Duration") %>%
mutate(variable = gsub("_", " ", variable))
figure <- ggplot(data_long, aes(x = Duration, y = value)) +
geom_line(color = '#737373') +
theme_minimal() +
facet_wrap(~ variable, scales = "free_y", ncol = 3) +
labs(title = title,
y = NULL) +
theme(plot.title = element_text(hjust = 0.5))
print(figure)
}The first six motion parameters are the standard motion parameters and include:
# load 6-rigid body motion parameters
# add duration
motion_file <- "data/data_demeaned.csv"
motion_demeaned <- read_csv(motion_file, col_names = F) %>%
mutate(Duration = c(1:nrow(.)))
# rename columns
motion_variables <- c(paste0("Translation_", c("X", "Y", "Z")),
paste0("Rotation_", c("X", "Y", "Z")))
colnames(motion_demeaned) <- c(motion_variables, "Duration")
# graph motion parameters
graph_motion(motion_demeaned, "6-Rigid Body Motion Parameters")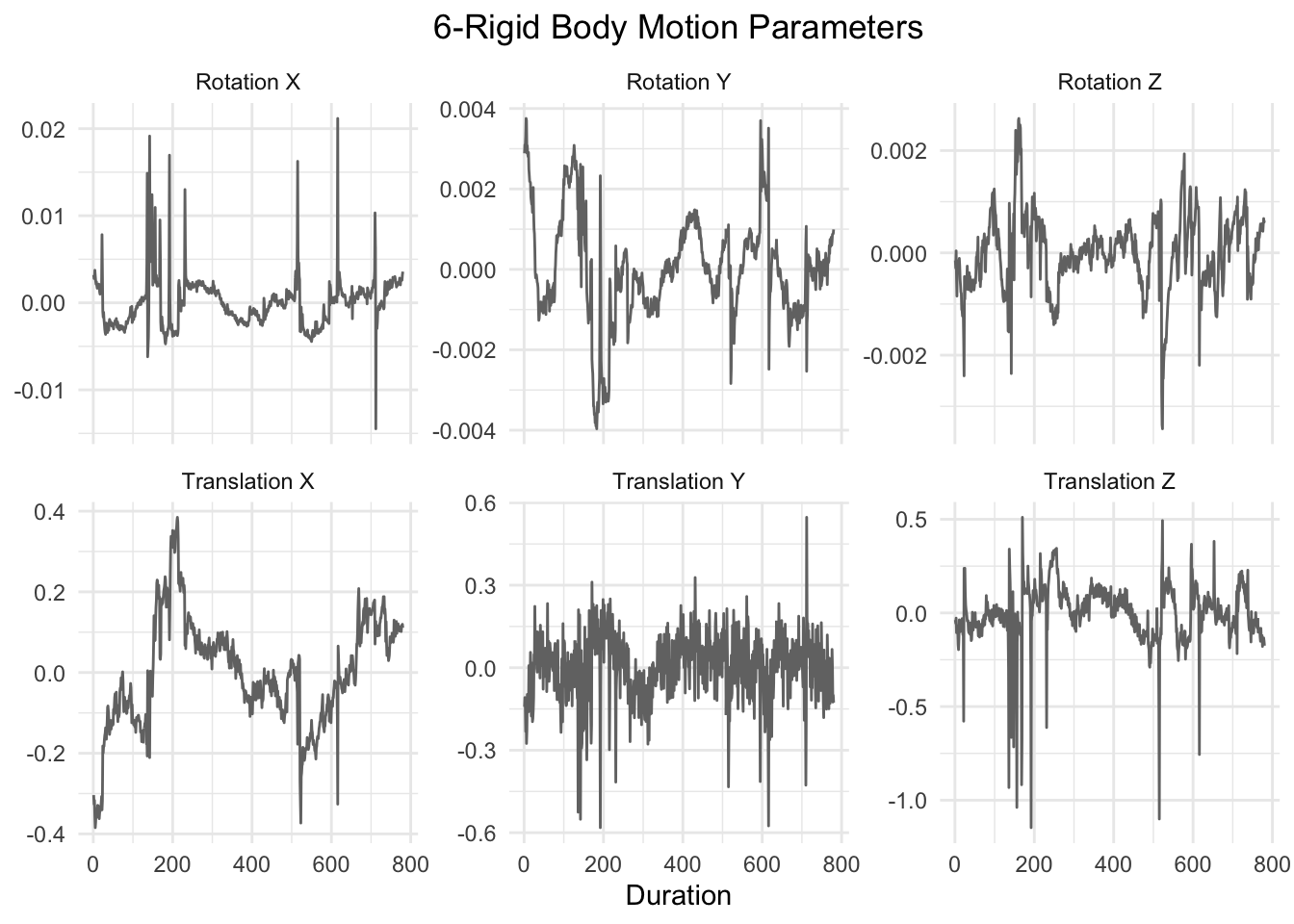
The next six motion parameters are the first temporal derivatives of the original six rigid body motion parameters. Specifically, it is the motion minus the motion from the previous time point.
# calculate first temporal derivative
motion_demeaned_td1 <- motion_demeaned %>%
sapply(., FUN = function(x) c(NA, diff(x))) %>%
as_tibble() %>%
mutate(Duration = c(1:nrow(.)))
# rename columns
colnames(motion_demeaned_td1) <- c(paste0(motion_variables, "_TD1"), "Duration")
# graph motion parameters
graph_motion(motion_demeaned_td1, "First Temporal Derivatives of the 6-Rigid Body Motion Parameters")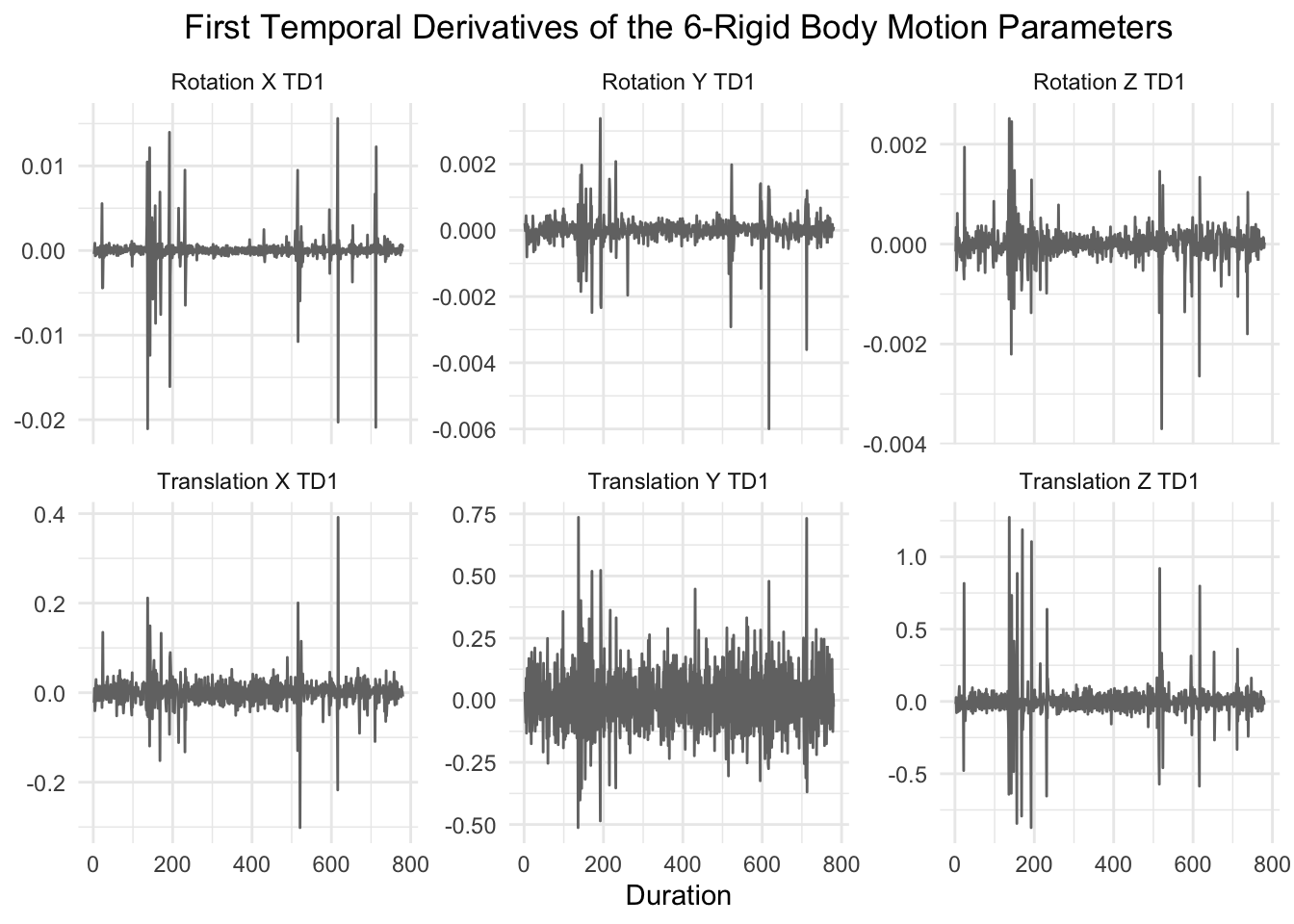
The next six motion parameters are the original six rigid body motion parameters squared:
# square 6-rigid body motion parameters
motion_demeaned_sq <- motion_demeaned^2 %>%
as_tibble() %>%
mutate(Duration = c(1:nrow(.)))
# rename columns
colnames(motion_demeaned_sq) <- c(paste0(motion_variables, "_Squared"), "Duration")
# graph motion parameters
graph_motion(motion_demeaned_sq, "6-Rigid Body Motion Parameters Squared")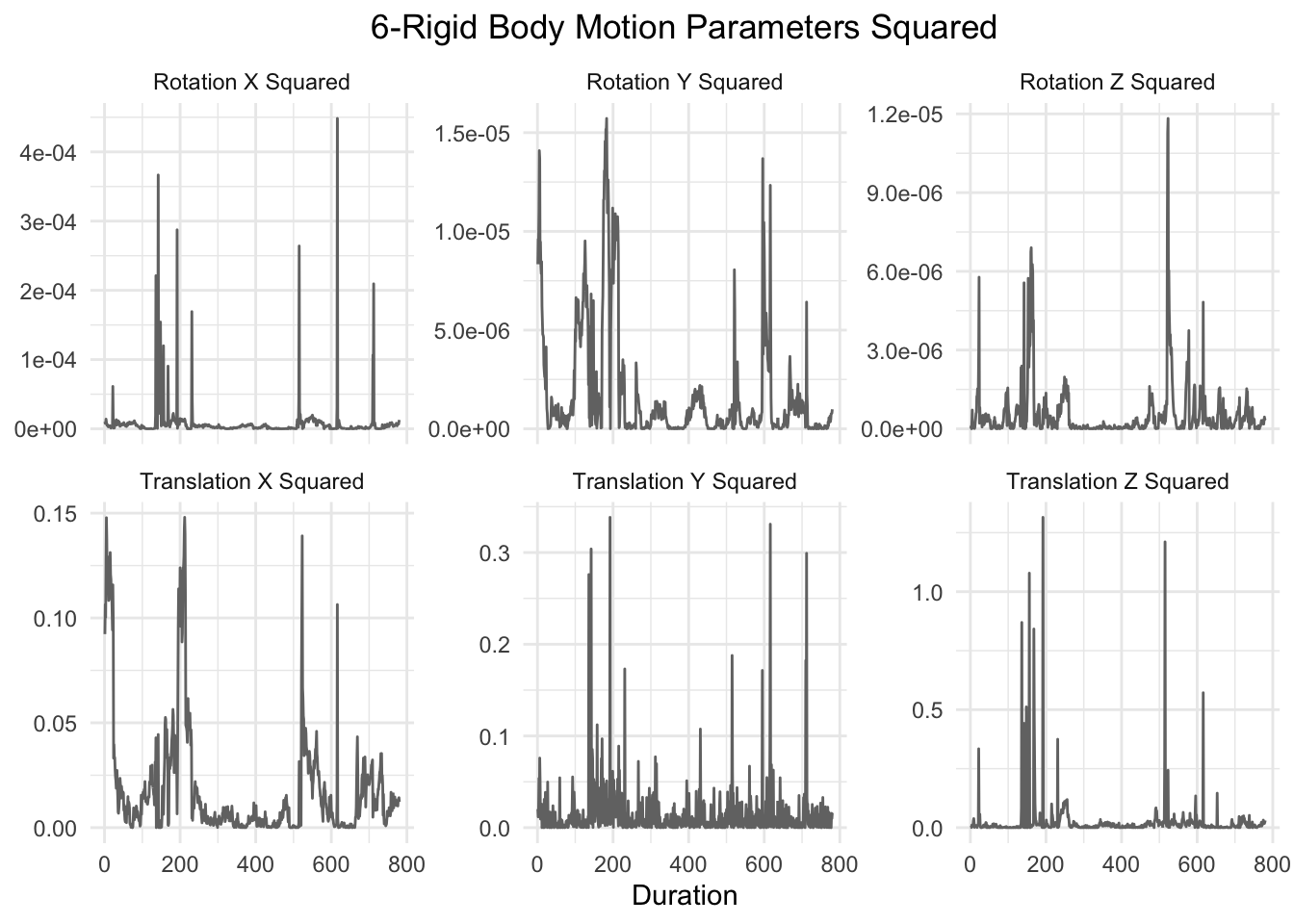
The next six motion parameters are the first temporal derivatives squared.
# square the first temporal derivative
motion_demeaned_td1_sq <- motion_demeaned_td1^2 %>%
as_tibble() %>%
mutate(Duration = 1:nrow(.))
# rename columns
colnames(motion_demeaned_td1_sq) <- c(paste0(motion_variables, "_TD1_Squared"), "Duration")
# plot motion.td1.sq into one figure
graph_motion(motion_demeaned_td1_sq, "First Temporal Derivatives of the 6-Rigid Body Motion Parameters Squared")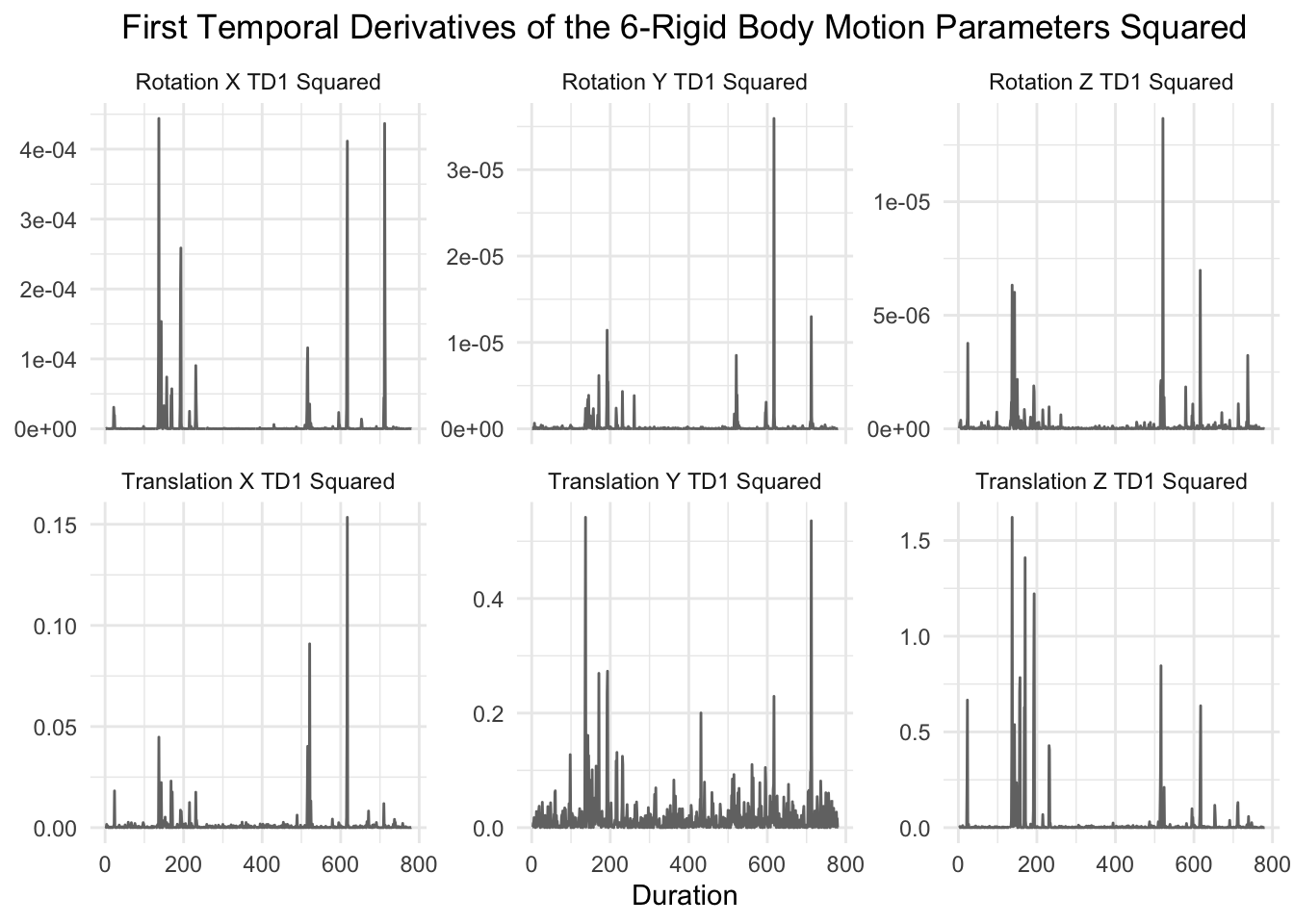
# function to create 24 motion variables =======================================
create_24_motion_variables <- function(dataset) {
# mean-center (demean) ----
# subtract each score from its respective column mean
dataset_motion_demeaned <- scale(x = dataset, center = T, scale = F)
# first temporal derivatives ----
# subtract motion from prior motion time point
dataset_motion_td1 <- sapply(dataset_motion_demeaned, FUN = function(x) c(NA, diff(x)))
# combine the variables into one dataset
dataset_motion <- data.frame(dataset_motion_demeaned, dataset_motion_td1)
# squares ----
# square 6-rigid body motion parameters and its temporal derivatives
dataset_motion_squared <- dataset_motion^2
# combine the variables into one dataset
dataset_motion <- data.frame(dataset_motion, dataset_motion_squared)
return(dataset_motion)
}
# load 6-rigid body motion parameters file =====================================
# assign to dataset_motion
dataset_motion <- read_csv(file.choose())
# create 24 motion variables using function ====================================
# assign output to dataset_motion_24
dataset_motion_24 <- create_24_motion_variables(dataset = dataset_motion)
# rename columns ===============================================================
# assign original motion variables
motion_variables <- c(
"trans_x", "trans_y", "trans_z",
"rot_x", "rot_y", "rot_z"
)
# rename columns
colnames(dataset_motion_24) <- c(
motion_variables,
paste0(motion_variables, "_td1"),
paste0(motion_variables, "_sq"),
paste0(motion_variables, "_td1_sq")
)
# export 24-motion variables ===================================================
write_csv(x = dataset_motion_24, file = "dataset-motion-24.csv")Note: Similar logic can also be applied to cerebral spinal fluid (CSF) and white matter (WM) time series to obtain an additional 8 nuisance regressors (32 nuisance regressors in total).
Friston, K. J., Williams, S., Howard, R., Frackowiak, R. S. J., & Turner, R. (1996). Movement-related effects in fMRI time-series. Magnetic Resonance in Medicine, 35(3), 346–355. http://doi.org/10.1002/mrm.1910350312
Yan, C.-G., Cheung, B., Kelly, C., Colcombe, S., Craddock, R. C., Di Martino, A., … Milham, M. P. (2013). A comprehensive assessment of regional variation in the impact of head micromovements on functional connectomics. NeuroImage, 76, 183–201. http://doi.org/10.1016/j.neuroimage.2013.03.004